3dsvm - an SVM-Light plugin for AFNI
Description
3dsvm is a command-line program and plugin for AFNI, built around SVM-Light. It provides the ability to analyze functional magnetic resonance imaging (fMRI) data as described in (LaConte et al., 2005).
Features:
- Reading AFNI-supported binary image formats including NIfTI
- Masking of variables (brain pixels)
- Censoring training samples
- Visualizing alphas as time series and linear weight vectors as functional overlays
- Classifying multiple categories
- Using non-linear kernels
- Performing classification and regression
- Performing real-time analysis for neurofeedback
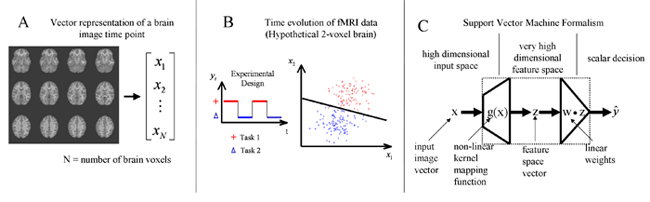
SVM brain state classification: (A) Each image is represented as an N voxel-dimensional vector. (B) A block design experiment and intensity changes for two voxels. Based on the distribution of intensities, a classification model (decision line) can be estimated. After the model is determined, it can be used to assign a brain state to new images. (C) Illustration of the SVM. The dotted box represents any (general) classifier. Inside the dotted box is a conceptual representation of the SVM approach.
AFNI began in the mid 1990's and is still an actively developing project written primarily in C (requires Unix, X11, and Motif). It provides great flexibility to the user through both the Unix shell and a gui interface. AFNI is used world-wide, open-source, maintained and developed by the Scientific and Statistical Computing Core at the National Institute of Mental Health (NIH, DHHS, USA, Earth), and provides great flexibility to the user through both the Unix shell and a gui interface.
SVM-Light is the core of 3dsvm. To be more compatible with fMRI data, 3dsvm reads AFNI-supported binary image formats, including the NIfTI format. It also allows for region of interest (ROI) 'masking' of variables (brain pixels), discarding training samples, and visualizing alphas and linear weight vectors. It can also handle multiple category classes. Currently 3dsvm uses SVM-Light V5.00.
We would like to thank Thorsten Joachims for providing SVM-Light to the scientific community, and for his enthusiastic support of this project. Similarly we would like to thank Bob Cox for AFNI and it's plugin support and Ziad Saad for his amazing support. Thanks also to Giuseppe Pagnoni and Stephen Strother for their comments and advice and Kyle Simmons and Michael Beauchamp for testing early versions of the plugin.
3dsvm is an active project in the LaConte Lab at the Virginia Tech Carilion Research Institute in Roanoke, VA (see developers). Stephen LaConte and Jeffrey Prescott developed early versions of 3dsvm at the Biomedical Imaging Technology Center (Xiaoping Hu, director) Department of Biomedical Engineering at Georgia Tech and Emory University. This initial work was supported by the National Institute of Health (NIH) National Institute of Neurological Disorders and Stroke (NINDS) R21NS050183.
Graphical User Interface
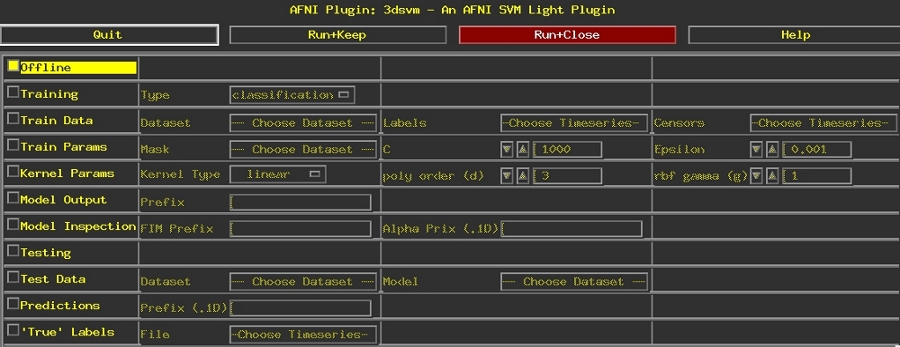
A screen shot of the graphical user interface for the 3dsvm plugin is shown below. It can be accessed trough the afni controller by selecting: Define Datamode ➜ Plugins ➜ 3dsvm.
Move your mouse over regions of the interface for further description.
Data & Use
Usage:
------
3dsvm [options]
Examples:
---------
1. Training: basic options require a training run, category (class) labels
for each timepoint, and an output model. In general, it usually makes
sense to include a mask file to exclude at least non-brain voxels
3dsvm -trainvol run1+orig \
-trainlabels run1_categories.1D \
-mask mask+orig \
-model model_run1
2. Training: obtain model alphas (a_run1.1D) and
model weights (fim: run1_fim+orig)
3dsvm -alpha a_run1 \
-trainvol run1+orig \
-trainlabels run1_categories.1D \
-mask mask+orig \
-model model_run1
-bucket run1_fim
3. Training: exclude some time points using a censor file
3dsvm -alpha a_run1 \
-trainvol run1+orig \
-trainlabels run1_categories.1D \
-censor censor.1D \
-mask mask+orig \
-model model_run1
-bucket run1_fim
4. Training: control svm model complexity (C value)
3dsvm -c 100.0 \
-alpha a_run1 \
-trainvol run1+orig \
-trainlabels run1_categories.1D \
-censor censor.1D \
-mask mask+orig \
-model model_run1
-bucket run1_fim
5. Training: using a kernel
3dsvm -c 100.0 \
-kernel polynomial -d 2 \
-alpha a_run1 \
-trainvol run1+orig \
-trainlabels run1_categories.1D \
-censor censor.1D \
-mask mask+orig \
-model model_run1
6. Training: using regression
3dsvm -type regression \
-c 100.0 \
-w 0.001 \
-alpha a_run1 \
-trainvol run1+orig \
-trainlabels run1_categories.1D \
-censor censor.1D \
-mask mask+orig \
-model model_run1
7. Testing: basic options require a testing run, a model, and an output
predictions file
3dsvm -testvol run2+orig \
-model model_run1+orig \
-predictions pred2_model1
8. Testing: compare predictions with 'truth'
3dsvm -testvol run2+orig \
-model model_run1+orig \
-testlabels run2_categories.1D \
-predictions pred2_model1
9. Testing: use -classout to output integer thresholded class predictions
(rather than continuous valued output)
3dsvm -classout \
-testvol run2+orig \
-model model_run1+orig \
-testlabels run2_categories.1D \
-predictions pred2_model1
Real-time Capabilities
The 3dsvm plugin has been integrated into AFNI's real-time framework and can be used for SVM-based analysis of fMRI data and neurofeedback. It takes advantage of existing real-time features (such as motion tracking and correction, running correlation analysis and TCP/IP communication) in AFNI. Take a look at this 2011 OHBM poster for more details.
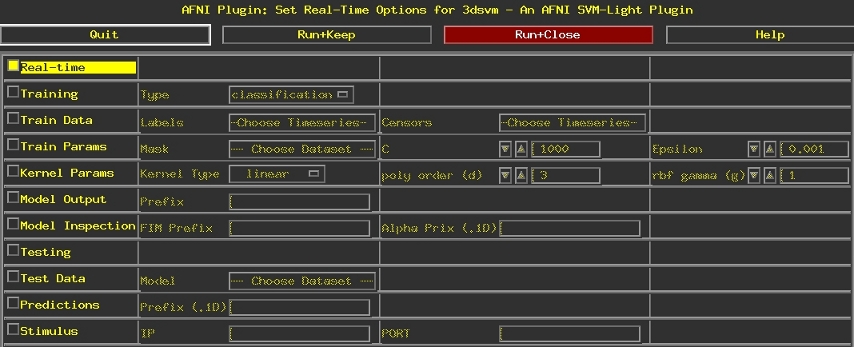
The graphical user interface illustrated below can be used to configure the real-time options of 3dsvm. It can be accessed trough the afni controller (started with the -rt option, i.e., afni -rt) by selecting: Define Datamode ➜ Plugins ➜ RT 3dsvm.
Move your mouse over regions of the interface for further description.
The real-time setup of the 3dsvm plugin can also be automated using AFNI's plugout_drive command. Using plugout_drive to set up the 3dsvm plugin for real-time experiments is very similar to the usage of the command-line program 3dsvm for off-line SVM analysis. Most of the 3dsvm (and SVM-Light) command-line options can be used in conjunction with plugout_drive.
Usage:
------
plugout_drive -com '3DSVM [options]'
Examples:
---------
Training:
plugout_drive -com '3DSVM -rt_train -trainlabels run1_categories.1D ...
-mask mask+orig -model model_run1'
Testing:
plugout_drive -com '3DSVM -rt_test -model model_run1+orig ...
-stim_ip 111.222.333.444 -stim_port 5000'
N.B.: -rt_train and -rt_test serve as flags for the real-time training
and testing modes, respectively. No brik or nifti file is
specified since it is expected from the scanner (or rtfeedme).
Download info
3dsvm is distributed with AFNI. You can go directly to the AFNI download page.
Developers
Current:
| Stephen M. LaConte | |
| Jonathan Lisinski | |
| You? Help us catch bugs and extend 3dsvm! |
Former:
| Jeff W. Presscott | |
| Will Curtis | |
| Jason White | |
| Ziad Saad |
References
When citing 3dsvm, please use the following reference:
LaConte, S., Strother, S., Cherkassky, V. and Hu, X. 2005. Support vector machines for temporal classification of block design fMRI data. NeuroImage, 26, 317-329.
Please also consider referencing:
T. Joachims, Making Large-Scale SVM Learning Practical.
Advances in Kernel Methods - Support Vector Learning,
B. Schoelkopf and C. Burges and A. Smola (ed.), MIT Press, 1999.
RW Cox. AFNI: Software for analysis and visualization of
functional magnetic resonance neuroimages.
Computers and Biomedical Research, 29:162-173, 1996.